Introduction
It is widely recognized that many small molecule drugs exhibit polypharmacology, whereby a single drug can affect multiple (protein) targets, resulting in off-target or undesired side effects. Identifying the protein targets or understanding drug mechanism of action (MOA) — how these small molecule compounds act to induce pharmacologic changes, however, have remained elusive. In a new tour de force paper, Mitchell et al. employed a high thoroughput proteomics workflow years in the making to help decipher the MOAs of 875 compounds.
In 2015, Chernobrovkin et al. demonstrated with FITExP the ability to filter out cell death noise to reveal known drug targets for six anticancer compounds using mass spectrometry (MS)-based proteome data alone. This drug MOA deconvolution method was replicated in multiple cell lines and expanded beyond cancer compounds by Saei et al. and scientists at Merck.
Polypharmacology widely observed and only a minority of ‘protein targets’ are regulated by their respective compounds
In a large effort that exceeds the throughput of past published experiments by tenfold, Mitchell et al. profiled proteome remodeling (change in protein abundance) after incubating 875 pharmacological compounds (targeting 914 proteins) in colon cancer cells (HCT116) for 24 hrs. After cell lysis, tryptic digestion, and TMT11-plex labeling, they measured more than 11 million peptide events using MS. A regulation threshold of more than two-fold change (compared to control) or greater than five standard of deviation, based on the protein abundance changes in all compounds assayed, was set. The regulation effects were largely replicated in a second set of biological replicate experiment performed at least six months later.
Mitchell et al. were able to identify among regulated proteins known compound targets (eg. pomalidomide) and found that only ~15% of compounds regulate their protein targets. The last observation was not surprising considering similar rate (25%) was detected from a prior proteomics study. In general, the direction of compound-induced protein change tend to skew toward one direction (up- or down-regulated), with a wide range of regulation effects from zero to more than two thousand proteins. The median compound regulation was fifteen proteins.
Compound perturbed proteome data revealed MOAs and off-target effects
Using this rich resource of deep proteome depth (median of 8,763 proteins per compound), Mitchell et al. found similar regulated proteins among compounds with same targets (e.g. BRD4 inhibitors). Further, they propose new targets for other compounds (eg. DHFR for three new compounds: imiquimod, R848, and ZD7288) based on similar proteome fingerprint.
As expected, the construction of compound-compound network found correlation among compounds with similar targets (e.g. nutlin-based MDM2 inhibitors, proteosome inhibitors). Moreover, they were able to identify common MOA of iron chelation among three highly correlated aroylhydrazones (SP2506, PAC-1, and KH7), each ascribed as inhibitors of different proteins targets LSD1, CASP3, and ADCY10, respectively.
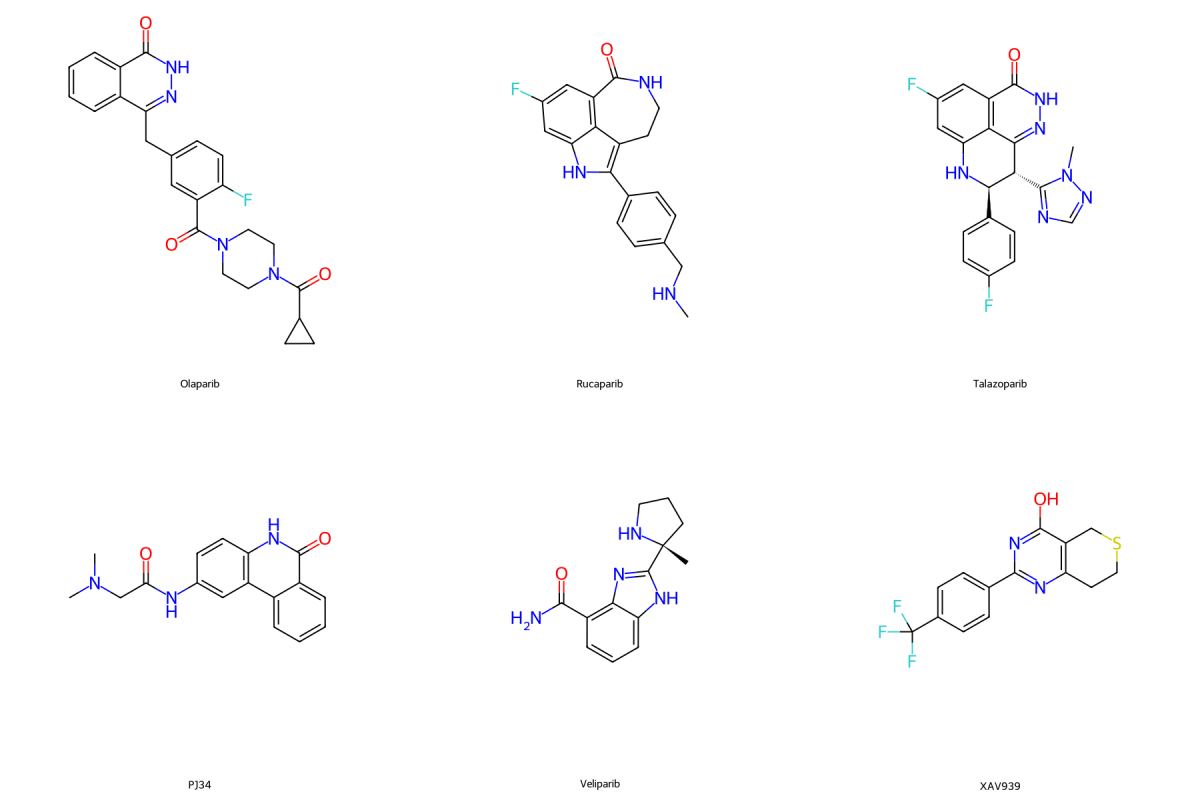
Comparing proteome differences between compounds that share the same targets revealed off-target effects. Of the two FDA approved poly (ADP-ribose) polymerase (PARP) inhibitors, talazoparib and olaparib, only talazoparib showed dose-dependent off-target effects.
Similarly, despite the lack of Bruton’s tyrosine kinase (BTK) expression in HCT116 cells, two BTK inhibitors: FDA-approved ibrutinib, and investigational RN486, also displayed divergent proteome changes. Follow-up experiments with a protein target deconvolution assay, in-cell proteome integral solubility alteration assay (PISA), showed that RN486 did not alter the thermal stability of kinases (which ibrutinib did) but instead altered the thermal stability in both ER and mitochondrial proteins. RN486 is not currently being studied in clinical trials.
JP1302 is an RNA transcription inhibitor whose MOA include linker histone H1
Lastly, to demonstrate the full utility of their resource, Mitchell et al. conducted several follow-up experiments based on their proteome data to deconvolute the MOA of a poorly characterized a tool compound JP1302 as an RNA transcription inhibitor. Like RN486, JP1302’s annotated protein target of alpha-adrenergic receptor antagonist (ADRA2C), is not detected in HCT116 cells. However, JP1302’s proteome fingerprint correlation with several RNA transcription inhibitors including CBL0137, a FACT complex inhibitor, yielded MOA insights toward RNA transcription inhibition and linker histone H1 degradation. These were confirmed by additional experiments, including in-cell PISA.
Proteomics in drug MOA deconvolution
High throughput proteomic technologies have lagged behind advances in transcriptomics due to higher expense and technological challenges. Mitchell et al. have addressed some of the technological challenges and demonstrated the utility of MS-based proteomics alone to deconvolute MOAs of small molecules. Standard experimental caveats including cell line, compound concentration (and/or toxicity), and reproducibility (correlation of ibrutinib to a repurchased ibrutinib was ~0.6 in this study) apply to this study. Given that over 90% of all drugs target proteins, this powerful resource can complement other existing resources (e.g. Connectivity Map, BioPlex) to help construct compound-induced cellular landscape toward more efficient drug development.
For help extracting insights from this resource and/or combining with other datasets, reach out at blindspotbio.
Special Offer
Free Bioinformatics or Research Consultation
Mention BLOG for free 20 min bioinformatics or research services help!
Schedule a ConsultationDive Deeper
- Commentary in Nature Reviews Drug Discovery
- Paper: A proteome-wide atlas of drug mechanism of action DOI
- Companion website to the paper
- Processed (paper supplementary data) data including proteome fingerprints and correlation tables.
- Unprocessed MS (.RAW) data at MassiVE: MSV000089282